by Alvin Kuriakose Thomas, Nadine Schmieder-Galfe, Yixin Zhang
The matrisome is an inventory of extracellular matrix proteins. The core matrisome comprises of about 300 proteins in mammals, in addition to matrix modifying enzymes, growth factors, and other associated proteins. Interactions of the various proteins in the extracellular environment provides the physical and chemical stimuli for biological responses. The modular, yet diverse nature of the extracellular biomolecules reflects their role in evolution, templating the transition to multicellularity and further on.1
Cataloging the essential building blocks of cell scaffolds and the parameters involved in the cell-environment interactions requires extensive data collection and analysis through trial and error experiments – an expensive and lengthy process. Alternatively, basic principles of the complex, multifactorial and interdependent cell-scaffold interactions could be established by parallel testing on a modular platform. A screening approach would enable the identification of the working principles to develop the right materials for the biomimicry of the extracellular environments and subsequent therapies.2
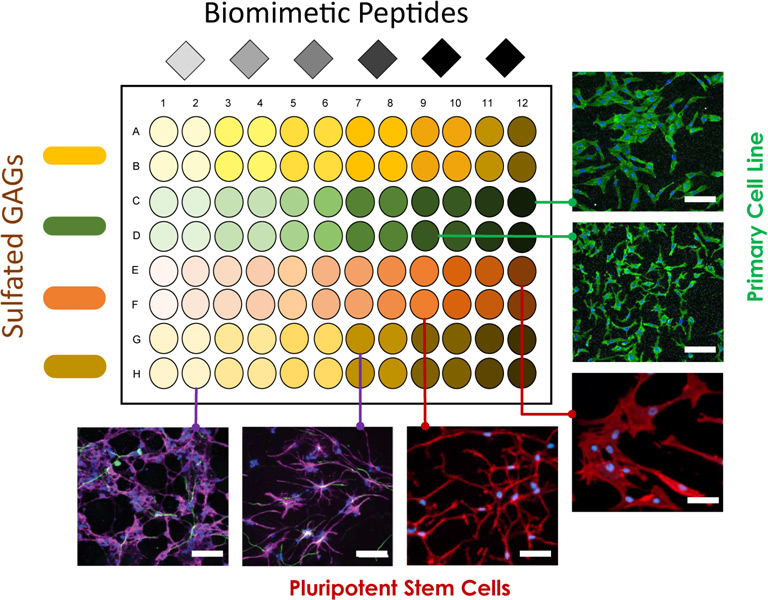
The non-covalent hydrogel technology at denovoMATRIX [*] based on crosslinking glycosaminoglycans simplifies the assembly of diverse combinatorial libraries of biomaterials, thus enabling efficient high throughput biomimetic material screening. The biologically relevant and modular hydrogel could comprise of functional synthetic peptides derived from natural extracellular proteins such as collagen, fibronectin, laminins, and growth factors etc. crosslinked to various glycosaminoglycans. Pilot screening studies have been successfully completed for several pluripotent stem cells, in addition to different primary, immortalized and cancer cell lines. The key scientific advantage is that cells can be studied in a biologically relevant environment compared to the studies on plastic or protein/peptide coated surfaces. Since, the material is transparent and coated onto standard cell-culture ware, routine cell culture protocols and analysis procedures based on histochemistry, microscopic analysis, etc. can be easily performed.3
The defined materials provided reproducible results, reliable and transferable for clinical applications.4
- Hynes, R. O.; Naba, A. Overview of the Matrisome-An Inventory of Extracellular Matrix Constituents and Functions. Cold Spring Harb. Perspect. Biol. 2012, 4 (1).
- Ahmed, M. Screening of 3D Biomaterials- A New Dimension in Biomaterials Research (or Two?). J. Tissue Sci. Eng. 2012, 3 (4), 3–4.
- Wieduwild, R.; Tsurkan, M.; Chwalek, K.; Murawala, P.; Nowak, M.; Freudenberg, U.; Neinhuis, C.; Werner, C.; Zhang, Y. Minimal Peptide Motif for Non-Covalent Peptide-Heparin Hydrogels. J. Am. Chem. Soc. 2013, 135 (8), 2919–2922.
- Tondera, C.; Wieduwild, R.; Röder, E.; Werner, C.; Zhang, Y.; Pietzsch, J. In Vivo Examination of an Injectable Hydrogel System Crosslinked by Peptide-Oligosaccharide Interaction in Immunocompetent Nude Mice. Adv. Funct. Mater. 2017, 27 (15), 1605189.

Combinatorial Biomaterial Screening is a promising strategy to discover biomaterials for tailored cell culture applications. Biomaterial libraries are generated using combinations of sulfated glycosaminoglycans and modified peptides. The library presents simultaneously relevant biochemical, physical and mechanical features of the extracellular matrix. Screening these biomaterials has revealed preferred biomatrices various cell types, including primary mesenchymal stromal cells (MSC) and primary neural precursor cells (NPC). Incorporation of GAGs sustains the expansion of all tested cell types comparable to existing cell culture surfaces, while osteogenic differentiation of MSC and neuronal differentiation of NPC were promoted certain biomatrices, respectively. The presented non-covalent system provides a powerful tool for developing tissue-specific ECM mimics. Scale bar: 50 µm.
picture credits: Robert Wieduwild, Richard Wetzel and Alvin Thomas
[*] denovoMATRIX – EXIST-Forschungstransfer Project at the Zhang Lab,
B CUBE Center for Molecular Bioengineering, Technische Universität Dresden